4.1. Deoxyribonucleic acid (DNA) and deoxyribose
DNA is a type of nucleic acid and is a substance that mainly stores genetic information of living things in the nucleus of a cell. The main function of DNA is to store information over a long period of time. The structure of DNA appears to be a helical structure as a whole, which corresponds to a skeleton and consists of nucleobases between the skeletons, all of which are linked by covalent bonds. The backbone has a long chain-like shape with a phosphate group bonded to deoxyribose, a monosaccharide. There are two types of bases: purine bases and pyrimidine bases. Purine bases include adenine and guanine, and pyrimidine bases include thymine and uracil. DNA can maintain a double helix structure because adenine bonds with thymine and guanine with cytosine hydrogen bonds.
Deoxyribose is a type of monosaccharide that lacks one oxygen atom in ribose. Deoxyribose is a component of DNA and belongs to a nucleotide unit and is covalently bonded to a phosphate group and a base, respectively. Each carbon atom in deoxyribose is numbered, starting from the rightmost carbon and clockwise from carbon 1 to carbon 5, as shown in the figure below.

4.2. Biophysics
It is an attempt to understand the essence of life phenomena from the point of view of physical laws or matter by expanding and applying various methods of advanced physics to living things. For example, it is important to interpret how an organism's actions are made in view of the properties of the molecules or atoms that make up it. To understand this, the methods of physiology, biochemistry, and biology must be used comprehensively. Some important research achievements in biophysics include physical research on the structure and properties of molecules constituting living organisms, especially high molecular substances such as proteins and nucleic acids. This study was intended to study elasticity among the physical properties according to the structure of DNA.
4.3. TATA box
The TATA box is a DNA sequence found in the DNA promoter region of archaea and eukaryotes. It accounts for 24% of human genes.
The TATA box is a binding site for transcription factors or histones (binding of transcription factors blocks binding of histones and vice versa) and is considered to be a key region of the promoter sequence as it is involved in the transcription of RNA polymerase.
4.4. Molecular Dynamics
Molecular dynamics is a method to investigate the microscopic motion of individual molecules in solid, liquid, or gaseous states in detail. It is a technique to study the dynamic structure or physical properties of molecules by simulating molecular motion using a computer. In this study, the bending-modulus of DNA was studied based on molecular dynamics.
4.5. Program
4.5.1. Program for simulation
MD(Molecular Dynamics simulation)
It is a program that can know the coordinate change of DNA, and it can know the movement of molecules by using potential energy. If we set a specific environment in the MD program, input information about the molecular coordinates of DNA, information about potential energy, and run a simulation, we can know the movement of molecules constituting DNA in a specific environment.
Potential energy → force → mass → acceleration → position
VMD(Visual Molecular Dynamics)
VMD can be used to identify more general molecules because it can read VMD standard protein data bank (PDB) files and displays designed for the modeling, visualization and analysis of isoprotein, nucleic acid, lipid bilayer assemblies, and biological systems. Contained structure. It provides various methods such as simple dots and lines, CPK, cylinder, backbone tube and ribbons, cartoon and other VMD rendering. VMD can be used to analyze the trajectories of animation implementations and molecular dynamics (MD) simulations. In particular, VMD acts as a graphic representation for an external MD program by animating molecules that are simulated on a remote computer. In this study, after making DNA using VMD, it is put in a space made of only water (Water box) to create a PDB file.
NAMD
NAMD has won the Gordon Bell Award in 2002 and the Sydney FERNBACH Award in 2012, and is a design parallel molecular dynamics code for high-performance simulation of large biomolecular systems. Based on parallel objects, NAMD can scale hundreds of cores, with more than 50 cores for typical and largest simulations. It uses the molecular graphics program VMD for NAMD simulation setup and trajectory analysis - and also uses the file compatible AMBER, CHARMm and X-PLOR. In this study, the shape change over time is obtained using NAMD for the PDB file of DNA generated using VMD.
※ File format created in the process for simulation
- PDB(Protein Data Bank) File format, 단백질 정보 파일 형식은 텍스트로 된 파일 형식으로, 단백질 정보 은행에 실려있는 분자들의 3차원 구조를 설명하는 데 사용된다.
PSF File format: 분자들의 좌표계 정보를 저장하는 PDB File format을 토대로 각 원자 사이의 힘의 크기와 에너지 관계를 저장한다. - Conf File format: Simulation을 실행하기 위한 File format으로 PDB, PSF, inp file의 경로를 지정해 준다. 또한 Simulation의 환경(온도, 압력 등), Simulation할 시간과 원자 사이의 힘 계산방법에 대한 정보를 저장한다.
- inp File format: 원자 사이의 정전기적, 반데르 발스 상호작용에 관한 상수를 가지는 Parameter file이다.
- DCD File format: Simulation한 결과(원자들의 좌표)를 저장하는 file이다.
- PDB (Protein Data Bank) File format, protein information file format, is a text file format used to describe the three-dimensional structure of molecules in the protein information bank.
- PSF File format: Based on the PDB file format that stores the coordinate system information of molecules, the magnitude of force and energy relationship between each atom is stored.
Conf File format: It is a file format for executing the simulation, and the path of PDB, PSF, inp file is designated. It also stores information about the simulation environment (temperature, pressure, etc.), the time to simulate, and the method of calculating the force between atoms. - inp File format: Parameter file containing constants for electrostatic and van der Waals interactions between atoms.
- DCD File format: A file that stores the simulation result (coordinates of atoms).
4.5.2. Programs for data analysis
MATLAB
As a technical computing language, millions of engineers and scientists around the world use MATLAB to analyze and design systems and products. MATLAB is used in automotive active safety systems, interplanetary spacecraft, health monitoring devices, smart power grids, LTE cellular networks, etc. being used in the field. The MATLAB platform is optimized for solving engineering and scientific problems. The matrix-based MATLAB language is a way of expressing computational mathematics.
Data can be easily visualized and interpreted with built-in graphics. A variety of pre-built toolbox libraries are provided so we can get started right away with the algorithms our domain needs. The desktop environment makes it easy to experiment, explore, and discover. In this study, it is used to obtain the rotation matrix according to physical modeling, the probability distribution (P(w)) for torsion (w), and the bending-modulus of DNA.
VideoMach
The VideoMach function has the following functions. Supports video and image formats (MPEG, AVI, DivX, FLC, JPEG, PNG, TIF, GIF, BMP, etc.), audio format support (MP3, MP2, OGG, WAV, AC3), fast image sequence detection, 50,000 per second More than one image is supported. In this study, it is used to convert the result into an image after executing NAMD.
4.6. Prior research
4.6.1. DNA의 탄성에 관한 연구
음파를 통한 DNA탄성 연구
선행연구인 프랑스 그레노블에 위치한 ILL(Institut LaueLangevin)연구소의 물리학자들은 DNA가 얼마나 잘 음파를 전달하는 지를 측정함으로써 그 유연성(flexibility)을 조사하였다.(Physical Review Letters발표)
광집게를 이용한 DNA탄성 연구
광주과학기술원 기전공학과 나노시뮬레이션 연구실에서는 광집게를 통한 측정을 시도하였다. Ionic effects on the elasticity of single DNA molecules, Christoph Baumann 외 공저 에서는 광집게를 이용하여 lamda-박테리오파지 DNA가 들어 있는 용액의 이온에 따른 DNA의 지속 길이를 구하였다. 연구 결과 1 가 이온 용액의 DNA 지속 길이가 다 원자가 이온 용액 속의 DNA 지속 길이보다 크게 나온다.
선행연구와 본 연구의 차이점
The above studies measured the elasticity of DNA through an experimental method. Ionic effects on the elasticity of single DNA molecules is a study of the effect of ions on the elasticity of specific real DNA. In addition, it was not possible to study the elasticity of DNA according to the nucleotide sequence because it was not presented as a research method that can derive the elasticity of DNA according to the nucleotide sequence, which is the purpose of this study. Therefore, we designed an inquiry process that can quantitatively derive the elastic modulus of DNA according to the nucleotide sequence.
위의 연구들은 실험적인 방법을 통하여 DNA의 탄성을 측정하였다. Ionic effects on the elasticity of single DNA molecules 는 특정 실제 DNA의 탄성에 이온이 미치는 효과에 대한 연구이다. 또한 본 연구의 탐구목적인 염기서열에 따른 DNA의 탄성을 도출해 낼 수 있는 연구방법으로 제시된 것이 아니므로 염기서열에 따른 DNA의 탄성연구가 불가능하였다. 따라서 염기서열에 따른 DNA의 탄성계수를 정량적으로 도출할 수 있는 탐구과정을 설계하였다.
4.6.2. Helix의 탄성에 관한 연구
The elasticity of Helix was studied using WLC (Worm-like chain) model, and the physical analysis process is as follows. Among the coordinate systems created by any method, the neighboring coordinate systems are related by a rotation matrix and can be expressed as follows.
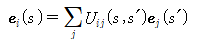
At this time, changes within the organized frame can be found through Frenet formulas.


: 반대칭 텐서

: 일반화된 비틀림
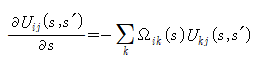
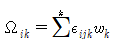
In the interval between s and s' divided by n, U in the above equation can be obtained.
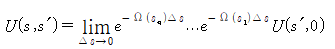
If linear elasticity exists, the elastic energy of the helix is E.
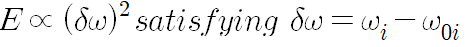
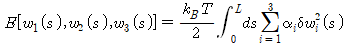
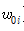
: 고유 비틀림의 집합
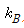
: 볼츠만 상수
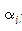
: 탄성계수(Bending-modulus)
Through the MD program, probability distributions P(w_1), P(w_2), P(w_3) for torsion(w) can be obtained. Interpreting the expression for , it can be seen that this distribution is a normal distribution. The value obtained by taking the negative sign of the natural logarithm of the obtained probability distribution is the same as the energy stored in the helix. At this time, the function for energy has the form of a quadratic function, and the elastic modulus (bending-modulus) can be obtained by matching the obtained quadratic function with the probability distribution function covered with the natural logarithm.
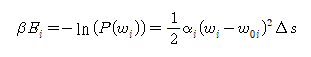
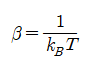



댓글